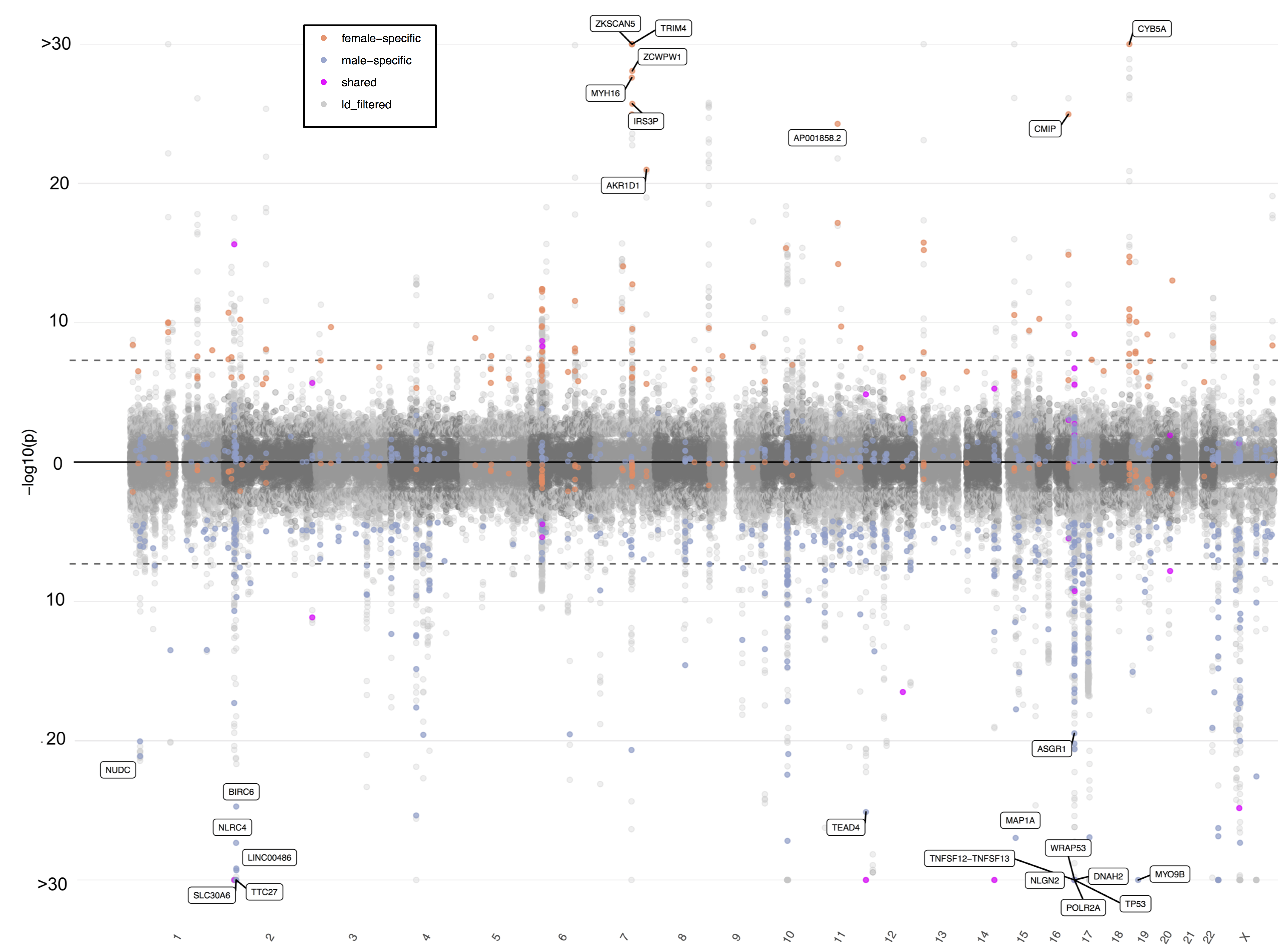
Sex-specific genetic effects across Biomarkers
This work was done in collaboration with Dr. Manuel Rivas and the Rivas lab.
Sex differences are prevalent in many clinical lab values, but the extent to which these differences are due to sex-differential genetic effects is unknown. We developed Sex Effects Mixture Model (SEMM), which is a Bayesian Mixture Model approach, for estimating sex-specific heritability, genetic correlation, and identifying sex-specific variants. We used sex-divided summary statistics from over 300,000 UK Biobank individuals. We found that the majority of the 33 biomarker traits examined did not show sex differences, with the exception of testosterone. For testosterone, we identified many variants with sex-specific effects.
This work was published in the European Journal of Human Genetics: Flynn, E., Tanigawa, Y., Rodriguez, F. et al. Sex-specific genetic effects across biomarkers. Eur J Hum Genet 29, 154–163 (2021). https://doi.org/10.1038/s41431-020-00712-w
SEMM is available as an R package here. This vignette demonstrates how the R package is used.
Additionally, the effect sizes of sex-specific testosterone variants are viewable in the Rivas Lab Global Biobank Engine: https://biobankengine.stanford.edu/sex-effects.
The code used for the publication is included in the repository: https://github.com/rivas-lab/sex-diff-biomarker-genetics.